Note
Click here to download the full example code
2.6.8.20. Cleaning segmentation with mathematical morphology¶
An example showing how to clean segmentation with mathematical morphology: removing small regions and holes.
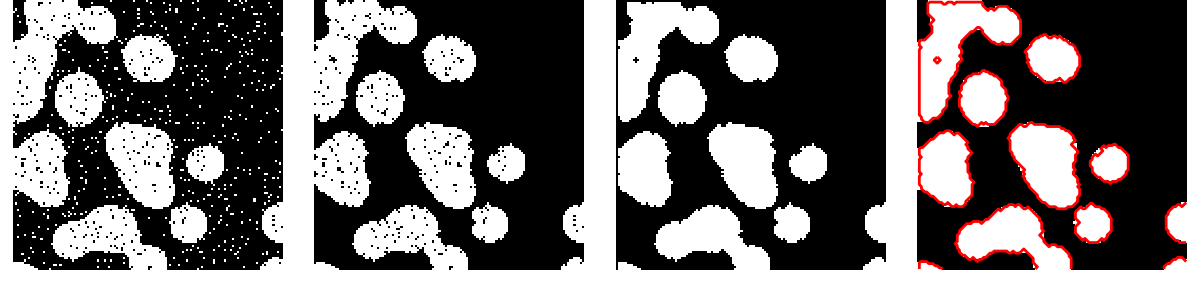
import numpy as np
from scipy import ndimage
import matplotlib.pyplot as plt
np.random.seed(1)
n = 10
l = 256
im = np.zeros((l, l))
points = l*np.random.random((2, n**2))
im[(points[0]).astype(np.int), (points[1]).astype(np.int)] = 1
im = ndimage.gaussian_filter(im, sigma=l/(4.*n))
mask = (im > im.mean()).astype(np.float)
img = mask + 0.3*np.random.randn(*mask.shape)
binary_img = img > 0.5
# Remove small white regions
open_img = ndimage.binary_opening(binary_img)
# Remove small black hole
close_img = ndimage.binary_closing(open_img)
plt.figure(figsize=(12, 3))
l = 128
plt.subplot(141)
plt.imshow(binary_img[:l, :l], cmap=plt.cm.gray)
plt.axis('off')
plt.subplot(142)
plt.imshow(open_img[:l, :l], cmap=plt.cm.gray)
plt.axis('off')
plt.subplot(143)
plt.imshow(close_img[:l, :l], cmap=plt.cm.gray)
plt.axis('off')
plt.subplot(144)
plt.imshow(mask[:l, :l], cmap=plt.cm.gray)
plt.contour(close_img[:l, :l], [0.5], linewidths=2, colors='r')
plt.axis('off')
plt.subplots_adjust(wspace=0.02, hspace=0.3, top=1, bottom=0.1, left=0, right=1)
plt.show()
Total running time of the script: ( 0 minutes 0.056 seconds)
